Genetic Characterization Of Hereditary, Sporadic, Dental Agenesis: Research On Pax9 And Msx1 Genes Mutation
Teresa Dinoi, Silvia Caruso, Paraskevi Ntrekou, PieraFiocchini, RobertoGatto, Giuseppe Marzo
University of L’Aquila, ViaVetoio 67100, Italia, L’Aquila, AQ, ITALY77054, United States
- *Corresponding Author:
- Silvia Caruso
University of L’Aquila, ViaVetoio 67100, Italia, L’Aquila
AQ, ITALY77054, United States
Tel: +393405363752
E-mail: silvia.caruso@graduate.univaq.it
Received date: 2015-07-23 Accepted date: 2015-08-25 Published date: 2015-09-02
Abstract
Objectives: The aim of this study is to find some genetic aspects of dental agenesis and, in particular, to analyze mutations in the MSX1 and PAX9 genes in individuals with non-syndromic dental agenesis.
The aim is to enhance our understanding of the genetic basis of these anomalies and to provide an epidemiological overview regarding the incidence of agenesis in the population, by innovative, minimally invasive, highly specific methods, following the rules of Good Clinical Practice (Good Clinical Practise, GCP) and in accordance with the Declaration of Helsinki.
Setting and Sample Population: Unit of Paediatric Dentistry of the Dental Clinic "Lidia Verza" of the Civil Hospital of Brescia. 26 patientes
Material and Methods: All patients were submitted to examination of the oral cavity, in order to identify the type of teeth, missing dental elements and any anomalies in the shape or in the structure of the elements in the dental arch. All patients were subjected to a blood sample of 10 cc of peripheral venous blood in order to perform the genetic analysis of PAX9 and MSX1.
Results: It is essential to remember that the research focused exclusively on the two genes PAX9 and MSX1. This means that other mutations in different genes possibly involved with a less obvious and more marginal role in the process of morphogenesis dental could be responsible for the agenesis phenotype found in the analyzed cases.
Conclusions: Considering anamnestic data, it has been supposed the kind of segregation of hereditary traits in family forms; possible ex novo mutations of these genes have not still been found for those who are supposed to have a sporadic form.
Introduction
A dental agenesis is a congenital lack of one or more dental elements. It basically is a failure of the enamel gum development, which causes the definitive absence of one or more elements [1] a frequent anomaly in the common population that plays an interesting role, both because the disorder is more frequently in the permanent dentition, and for the complexity of the therapeutic issues that causes.
Dental agenesis may be encountered more frequently as sporadic or familiar agenesis, in which deficit is normally limited to a no formation of one or more dental elements [1].
If the agenesis follow a syndromic pattern, they are configured as one of the effects that the disease causes on a systemic level; then they seem to be associated with other oral and dental anomalies [1].
Classification
The variations of these anomalies are better described by the classification suggested by Malavez [2], which defines:
• ANODONTIA: simultaneous absence of all teeth;
• OLIGODONTIA: presence of a number of teeth less than half of those normally present in the dental arches.
In order to classify the disease in relation to the affected sector, agenesis can be classified as follows [3].
• ANTERIOR: affecting the anterior elements;
• CENTRAL: localized between the first premolar and the first molar
• POSTERIOR: affecting the second molar and the third molar
• MIXED: when more than a single sector is involved.
Epidemiology
Dental agenesis is the most common development anomaly in humans4, the prevalence of the disease in general population, as reported in literature, varies from 1,6% to 11,3% [4-6].
In Italy, according to the studies conducted by [7] at least 5, 2% of population suffers from dental agenesis [7].
The prevalence in Italy rose to 9.1% in a recent study on 1190 patients by Luzzi et al. (2010) , referred to the unit clinical complex of Pediatric Dentistry at the University La Sapienza of Rome.
However, it should be pointed out that the percentage found in this study appears to be higher than that already verified in other studies, since the sample was obtained in a structure that offers a specialized, ortho-pedodontics service. This kind of service regards orthodontic and rehabilitation therapies, which are necessary for this type of anomalies and, therefore, the data refer to a specific population, selected for dental diseases. In fact, the percentage of patients with syndromic agenesis is 41.5%.
Females suffer of this disease with a ratio of 3:2 compared to males [3,8,9], however gender seems not to influence common agenesis patterns [9,10].
The most affected elements are the third molars, followed by the second lower premolars, upper lateral incisors and second premolars.
In fact [10] excluding third molars, upper lateral incisors and lower second premolars themselves account 85% of all congenitally missing teeth [11].
According to [3], distribution of agenesis in dental groups (excluding third molars) could be as follows:
• Lower second premolars: 38,6%
• Upper lateral incisors: 29,3%
• Upper second premolars: 16,5%
• Lower central incisors: 4%
• Second molars: 2,6%
• Canines: 2%
• Lower lateral incisors, upper central incisors, first premolars, first molars: 7%.
The unilateral agenesis are more common than the bilateral ones, except for the upper lateral incisors, which are most frequently absent simultaneously [5].
The overall prevalence of agenesis in the mandibular arch is similar to the maxillary arch, but there is a difference between the two dental arches considering the different dental elements [12].
Agenesis of deciduous dentition is found in literature with much lower values of prevalence, ranging between 0.08% and 1.55% [4-6].
Etiology
In the literature, several theories and hypotheses try to explain the causes of this disease.
For convenience, one can distinguish four different schools of thought:
• Evolutive/phylogenetic etiology
• Anatomical etiology
• Genetic etiology
The evolutive hypothesis regarding the etiology of agenesis is based on the theory of phylogenetic reduction of the dental formula. Agenesis is an adaptive mechanism, typical of the evolution of the species, in which the teeth are removed to adjust the number of teeth in the arch to the anterior-posterior decrease of the maxillo-mandibular complex and / or to the incorrect masticatory function due to diet alterations (soft and treated food) [13-15,20].
As far as anatomical theories are concerned, they pay particular attention to areas that are more easily affected by agenesis.
Svinhufvud et al. (1988) hypothesized the existence of areas of the dental lamina that, during the process of formation and maturation of the tooth, are more susceptible to the influences of epigenetic factors and therefore are more severely affected by dental anomalies such as agenesis.
Some examples are:
1. Agenesis of upper lateral incisors, which grow in the area of embryonal fusion between lateral maxillary processes and medial nasal processes;
2. Agenesis of lower second premolars which are developed in the distal area of primary dental lamina;
3. Agenesis of lower central incisors that originate in the median lamina, point of fusion of the two mandibular processes.
It is well known that both genetic and environmental factors may cause this disease [14].
There are numerous non-genetic, potential and verified factors that can lead to dental agenesis as they are able to arrest the development of affected dental elements by acting both directly (invasive) and indirectly [16].
Among these ones, in literature there are:
• Traumatic events [17].
• Antitumor therapies and local aneoplasia
• Infective processes
• Endocrine and metabolic pathologies
• Exposure to environmental toxines
• Taking medications during pregnancy
Due to intensive genetic studies in recent years and to the evidence provided by the hereditary patterns, it is now possible to confirm the genetic etiology as the most common and validated cause of agenesis.
Although the agenesis may occasionally be caused by environmental factors, in most cases the basis of this disease are to be found in genetic, most of all because it has been demonstrated that the entire dental development, in particular the position , the number, size and shape of the elements, is under strict genetic control.
Some factors that may indirectly indicate the important role of genetic factors in the etiology are:
1. The occurrence of cases among members of the same family, that is distributed over three generations (or at least two): agenesis is more common among patients that are related with affected individuals than the general population; (Brook AH., 1984)
2. The higher frequency of hypodontia in homozygotes than in heterozygotes;
3. The differences in prevalence in different populations and races;
4. The difficulty in finding one or more environmental factors responsible for the condition in each case of agenesis;
5. The association with inherited genetic syndromes (syndromic agenesis).
Genes involved in dental agenesis
There are several genes that are involved in dental agenesis (34 -35), mutations in genes MSX1, PAX9 have been demonstrated to be involved in the etiology of non-syndromic agenesis in humans, as described below.
MSX1
MSX1 is located on the short arm of chromosome 4 (4p16.2), it was the first gene to be definitively associated to dental agenesis in humans [18]. MSX1 mutations were generally related to hypodontia and oligodontia with autosomal dominant hereditary patterns: the second premolars and third molars are the most frequently affected teeth.
A nonsense mutation of the gene MSX1 was associated with dental agenesis and various combinations of lip / palatine cleft [19]. Recently it has been described a mutation of this gene that is responsible for autosomal recessive hypodontia [20].
PAX9
The PAX9 gene, located on the long arm of chromosome 14 (14q13.3), was the first to be associated with dental agenesis due to a frameshift mutation (insertion of a guanine that causes premature termination of protein) to nonsyndromic oligodontia with autosomal dominant hereditary pattern.
The observation of smaller teeth in affected individuals suggests that PAX9 is not only involved in the positioning and in the development of certain teeth, but also in the morphogenesis of the entire dentition [21,22].
Aim of the study
Advances in human genome project and in molecular genetics have led to substantial developments in the identification of genes involved in the pathogenesis of human diseases, including diseases that involve the formation of dental enamel and dentin and anomalies of number of teeth.
Genetic diseases that affect the tooth structure have been classified according to the affected tissue (enamel / dentin), their specificity (syndromic / non-syndromic) and their pattern of inheritance: autosomal dominant (AD), autosomal recessive (AR), or X-linked recessive (XLR).
Regarding dental agenesis it can be said that, finding in clinical practice the presence of a family history in patients with this disease, many scientific works have set themselves the goal of studying the genetic basis of agenesis, highlighting different patterns of transmission: autosomal dominant with incomplete penetrance and variable expressivity, autosomal recessive and X-linked gene (X-linked).
Nonsyndromic dental agenesis, either in familial forms and in sporadic forms, would involve mutations, inherited or ex novo, of one or more genes involved in the molecular processes of tooth development, such as, for example, PAX9, MSX1, EDA, AXIN2 and LTBP3.
The purpose of this study is to find some genetic aspects of dental agenesis and, in particular, to search for mutations in the genes PAX9 and MSX1 in individuals with non-syndromic tooth agenesis.
The kind of segregation of hereditary traits in family forms is then evaluated, while, in case of individuals, for whom a sporadic (non familiar) form is assumed, possible ex novo mutations in these genes are now been seeking.
Materials and Methods
Sample selection
This study has been approved as AOBS-GENI-2011, NP No. 1119 by the Ethics Committee of the ”Azienda Ospedaliera Spedali Civili” of Brescia. It took into account patients that referred to Unit of Paediatric Dentistry of the Dental Clinic "Lidia Verza" of the Civil Hospital of Brescia and that suffered from dental agenesis and had a particular familiarity with this disease or classified as involving sporadic agenesis. Patient selection has followed the following enrollment criteria.
• Inclusion Criteria:
- Presence of single or multiple dental agenesis;
- Familiarity with dental agenesis;
- Age between 6 and 18 years old;
- Subscription to informed consent
• Exclusion criteria:
- History of prior environmental factors or conditions which may have led to a lack of development of the dental follicle: trauma, cancer therapies, local tumor, infection, metabolic and endocrine disorders, exposure to environmental toxins, taking special drugs during pregnancy, fetal distress syndromes;
- No subscription to informed consent.
• Control choice:
After joining the informed consent, parents and brothers and sisters of patients, regardless the presence or absence of agenesis, were enrolled in the study as healthy controls.
The choice to include children in this study has been formulated on two key concepts:
- The possibility to track familiar pedigrees arriving at least to the second generation, essential prerogative for genetic studies;
- Possible difficulties in reconstructing the dental history in adults, due to poor memory of dental treatments, which is insufficient to exclude prior avulsions or dental trauma.
Examinations and investigations
Each patient enrolled in the study has been submitted, according to the operating protocol, the following clinical, radiographic and laboratory procedures. Part of the exams are part of standard clinical routine for this disease, such as physical examination, patient history and radiographic examination that, only together, allow to properly diagnose dental agenesis. Additional procedures are blood sampling and genetic analysis.
All patients were submitted to examination of the oral cavity, in order to identify the type of teeth, missing dental elements and any anomalies in the shape or structure of the elements in the dental arch.
The patients were subjected to an orthopantomographic test, as clinical routine during a first dental visit, in order to confirm the clinical picture of agenesis of teeth, to exclude the presence of included elements or possible intraosseous neo formations. All patients were subjected to a blood sample of 10 cc of peripheral venous blood in order to perform the genetic analysis of PAX9 and MSX1.
Blood sampling was performed at the Paediatric Dentistry Hospital of Spedali Civili of Brescia. The peripheral venous blood was collected in commercial test tubes containing anticoagulant lithium heparin, that has been immediately stored at 4°C until transfer, by specific thermal bag, at the Biology and Genetics Section (Dpt SBB) of the Faculty of Medicine of the University of Brescia.
Every test tube had an identification code that enabled to anonymize the sample and to guarantee the patient’s privacy and, at the same time, allowed the doctor to associate codes to patients.
This sperimental procedure aimed at discovering mutations due to PAX9 and MSX1 genes and it has been conducted at the Biology and Gentics Section (Dpt SBB) at the Faculty of Medicine of the University of Brescia.
After transferring the blood samples to the laboratory of genetics, these were extracted and evaluated for purity and quality of the genomic DNA and, in some cases, even the RNA, using methodical TRIzol® Reagent.
Then, DNA was used for sequencing the coding exonic regions and the parts between exon and intron to identify the presence of mutations in the chosen genes PAX9 and MSX1. Sequencing was done with BigDye® Terminator technology, using the automatic capillary sequentiator (Applied Biosystems®) in the laboratory.
Laboratory stage and genetic analysis
The spectrum of genetic mutations related to the development of non-syndromic agenesis in humans includes a large extent of MSX1 and PAX9 defects, encoding transcription factors that are fundamental in the development of teeth [4].
It has been demonstrated, by using knockout mice for PAX9 and MSX1, that the functional inactivation of these two genes involves stopping the process of odontogenesis and disorders in many other tissues.
These genes were chosen because of their key role in the process of odontogenesis, especially in the early stages: it is assumed, therefore, that an impairment caused by a possible genetic mutation may have consequences easily represented at the phenotypic level.
The biological material is genomic DNA from nucleated cells of the collected blood samples.
The genomic DNA was obtained by the TRIzol® Reagent method, through the application of protocols for extraction from samples of peripheral blood, using saline swabs.
The extracted DNA meets the requirements of stability over time and resistance to multiple freeze-thaw cycles. The method takes about a couple of days to be executed.
To determine the concentration of the extracted genomic DNA, a spectrophotometric measurement was performed by using the NanoDrop®, an instrument that allows obtaining a very accurate quantification.
This instrument is equipped with a software that processes the spectrophotometric reading and provides the concentration (ng / uL) of the test sample.
To use the PCR technique, it is necessary to set specific primers that allow amplifying only the segment of interest. For this study, specific primers were constructed by consulting the information available on the biological GenBank database.
Furthermore, it is appropriate to carry out tests to optimize operating conditions to draw up an operational protocol of use of PCR, specific for each gene of interest, allowing the unfolding of the amplification procedure in optimal conditions. The PAX9 gene has five exons. Six pairs of forward and reverse primers were used for the amplification of the four exons of primary interest (n ° 2, 3, 4 and 5) to detect possible mutations in the gene.
In fact, for the amplification of exon No. 2, which is particularly extended, it was necessary to use three pairs of primers to avoid the loss of information.
In particular for PAX9:
| PRIMER | Tm (°C) | Amplified Fragment (bp) | Sequence |
|---|---|---|---|
| 1F | 69,1 | 273 bp | 5’-GCCCACGTTGCTGCTTAGATTGAAA-3’ |
| 1R | 68,3 | 5’-CTCCCTCCCTTCCCGGCTCT-3’ | |
| 2aF | 71,3 | 235 bp | 5’-AGGCAGCTGTCCCAAGCAGCG-3’ |
| 2aR | 70,5 | 5’-TGTATCGCGCCAGGATCTTGCTG-3’ | |
| 2bF | 68,8 | 249 bp | 5’-ATCCGACCGTGTGACATCAGCC-3’ |
| 2bR | 67 | 5’-GGAGGGCACATTGTACTTGTCGC-3’ | |
| 2cF | 71,3 | 355 bp | 5’-GCATCTTCGCCTGGGAGATCCG-3’ |
| 2cR | 67,2 | 5’-GAGCCCCTACCTTGGTCGGTG-3’ | |
| 3F | 68,7 | 264 bp | 5’-TTTGGGTCCCGTCTCAAGAGTGG-3’ |
| 3R | 72,5 | 5’-CCTAAATCCCCGCCGCCACG-3’ | |
| 4F | 63 | 450 bp | 5’-GGAGAGTAGAGTCAGAGCATTGCTG-3’ |
| 4R | 65,6 | 5’-GAGACCTGGGAATTGGGGGA-3’ |
Amplification Condizions For Pax9
| n° cycles | DENATURATION | ANNEALING | EXTENSION |
|---|---|---|---|
| 1 | 5’ 95°C (hot start) | ||
| 35 | 20” 95°C | 20” 55°C | 20” 72°C |
| 1 | 3’ 72°C |
Instrument:
GeneAmp® PCR System 9700 (Applied Biosystems®)
The MSX1 gene has two exons. Three pairs of forward and reverse primers were used to examine both the genes.
In particular for MSX1
| PRIMER | Tm (°C) | Amplified Fragment (bp) | Sequence |
|---|---|---|---|
| 1aF | 64,3 | 420 bp | 5’-CCAGTGCTGCGGCAGAAG-3’ |
| 1aR | 59,7 | 5’-CTTCTGGCAGCTTGAGGAGT-3’ | |
| 1bF | 59,8 | 332 bp | 5’-AGTGTCCCCTTCGCTCCT-3’ |
| 1bR | 62,6 | 5’-GCCTGGGTTCTGGCTACTCA-3’ | |
| 2F | 60,1 | 664 bp | 5’-ACTTGGCGGCACTCAATATC-3’ |
| 2R | 59 | 5’-TGTGAGGGTTAAAGGGAAGG-3’ |
Amplification Conditions For MSX1:
| n° cycles | DENATURATION | ANNEALING | EXTENSION |
|---|---|---|---|
| 1 | 5’ 95°C (hot start) | ||
| 35 | 30” 95°C | 30” 60°C | 30” 72°C |
| 1 | 3’ 72°C |
Instrument:
GeneAmp® PCR System 9700 (Applied Biosystems®)
In short, the procedure is the following:
1. Denaturing DNA from double helix to single helix by incubation at 94°-95°C;
2. Cooling the solution and then allowing the pairing, i.e. the annealing, of primers between 37°C and 65°C, depending on the homology among the sequences of the bases of primers and the sequences of the bases of DNA. The two primers are meant to be paired on opposite DNA strands, at the ends of the sequence that has to be amplified. As a result, the 3’ ends of the primers face each other.
3. Extending the primer by using DNA polymerase at 70-75 ° C. A special heat-resistant DNA polymerase called Taq polymerase is used. This particular enzyme is the DNA polymerase of a thermophilic bacterium, the thermophilus aquaticus. This process is made possible due to an excess of available synthetic oligonucleotides.
4. Single stranded DNA and … in order to associate primers.
5. Repeating primers extention by Taq DNA polymerase. There are two double helix molecules: one has a unitary length (it has the same length as DNA between primer A’s 5’ end and primer B’s 5’ end); the other one in both the molecules is longer than the unitary length.
6. Repeating DNA denaturation and reassociating primers.
7. Repeating primer extention by Taq DNA polymerase in order to produce double helix DNA of unitary length. It is important to note that three cycles have been necessary to produce the molecules with the same length as the target DNA. Repeated cycles of denaturation, association and extension cause a geometric increase of DNA of unit length.
By using PCR, the amount of new DNA geometrically or exponentially increases: starting with a DNA molecule, a PCR cycle will produce two molecules, two cycles will produce four molecules, three cycles eight molecules, two of which are the target DNA.
Ten successive cycles will produce 1024 copies of the original DNA (210) and in 20 cycles there will be 1,048,576 copies (220) of the target DNA. The product of the polymerization chain is equal to 2n copies of template DNA: n is the number of amplification cycles and the substrate of each new cycle is the product of the extension reaction of the previous cycle.
The procedure is fast, since each cycle takes only a few minutes using a thermal cycler, a thermostatic machine that automatically performs temperature cycles of programmed temperature.
For each PCR reaction, a negative control without DNA was prepared, in order to identify possible contaminations.
A kit (Wizard® SV Gel and PCR Clean-Up System - Promega®) previously purified the amplification products obtained by genomic PCR.
For the sequence reactions, technology BigDye® Terminator was used, together with specific primers for each fragment. The peculiarity of this sequencing PCR is the incorporation of labeled nucleotides, each one with a different fluorescence (using 4 different fluorochromes) which will then be displayed during DNA sequencing.
The products of the sequence are then purified through a reaction of selective precipitation; in fact, to make a correct reading of the sequences, it is necessary to remove the unincorporated dideoxynucleotides.
The sequencing was carried out by using f the analyzer ABI PRISM® 3100 (Applied Biosystems®), which is based on an automatic system of capillary electrophoresis that evaluates the absorbance of the four different fluorochromes producing an electropherogram of the specific sequence.
Then, for analysis and file reading, specific bioinformatic programs were used, such as:
- NCBI (https://www.ncbi.nlm.nih.gov/)
- GeneCards (https://www.genecards.org/)
MutationDiscovery (https://www.mutationdiscovery.com/)
Results
This study involved 26 patients:
| Group | AFFECTED | NON AFFECTED |
|---|---|---|
| PROBAND | 15 | - |
| FAMILIARS | 5 | 6 |
For each patient, distinguished by numeric arbitrary cardinal code, was made a card for collecting data was made, indicating:
- Age at the time of the visit
- Sex
- detected agenesis (negativity, positivity and involved elements)
- OPT execution (yes / no)
- Presence of detected genetic mutations.
* A single nucleotide polymorphism
Among the 26 subjects who agreed to join the study, the genes MSX1 and PAX9 were sequenced for 9 patients.
| PILOT STUDY | |||
|---|---|---|---|
| PAX-9 | IDENTIFIED SEQUENCE VARIATIONS | MSX-1 | IDENTIFIED SEQUENCE VARIATIONS |
| Exon 2 | -- | Exon 1 | SNP* rs36059701 C/G |
| Exon 3 | -- | Intron1 | SNP rs1042484 T/C |
| Exon 4 | -- | Exon 2 | SNP rs8670 C/T |
| Exon 5 | -- | ||
Pilot Study
As a pilot study, which has de
veloped the protocol of the laboratory phase of the study, blood samples of the patient's parents were not collected, while, in retrospect, it was possible to reconstruct the pedigree of the case.
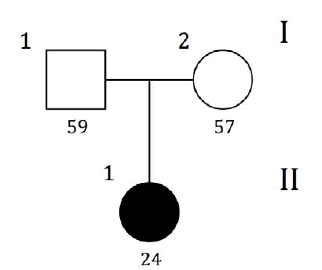
| GRAPHIC IDENTIFICATION | CASE | DATE OF BIRTH | AGENESIS CHARACTERISTICS |
|---|---|---|---|
| I;1 | - | 28/07/1953 | non affected |
| I;2 | - | 20/07/1955 | non affected |
| II;1 | PILOT | 05/09/1988 | 35,45 |
Sequencing Results
FAMILY n°1 (cases 1-4)
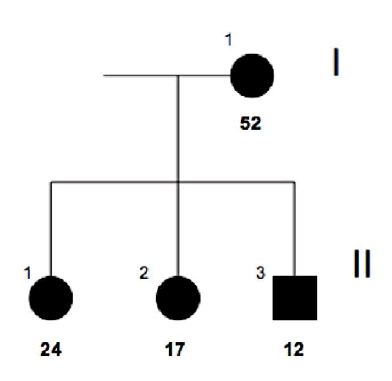
| GRAPHIC IDENTIFICATION | CASE | DATE OF BIRTH | AGENESIS CHARACTERISTICS |
|---|---|---|---|
| I;1 | 3 | 23/09/1960 | 15,25,35 |
| II;1 | 4 | 05/08/1988 | 35,45 |
| II;2 | 2 | 10/03/1995 | 12,22,34,41,44,47 |
| II;3 | 1 | 06/01/2000 | 11, 12, 13, 14, 21, 22, 23, 24, 31, 32, 33, 35, 41, 42, 43, 44, 45 |
Sequencing Results
| CASE 1 | |||
|---|---|---|---|
| PAX-9 | IDENTIFIED SEQUENCE VARIATIONS | MSX-1 | IDENTIFIED SEQUENCE VARIATIONS |
| Exon 2 | -- | Exon 1 | -- |
| Exon 3 | -- | Intron 1 | SNP rs1042484 T/C |
| Exon 4 | SNP rs12881240 C/T | Exon 2 | -- |
| Exon 5 | -- | ||
| CASE 2 | |||
|---|---|---|---|
| PAX-9 | IDENTIFIED SEQUENCE VARIATIONS | MSX-1 | IDENTIFIED SEQUENCE VARIATIONS |
| Exon 2 | -- | Exon 1 | -- |
| Exon 3 | -- | Intron 1 | SNP rs1042484 T/C |
| Exon 4 | -- | Exon 2 | -- |
| Exon 5 | -- | ||
| CASE 3 | |||
|---|---|---|---|
| PAX-9 | IDENTIFIED SEQUENCE VARIATIONS | MSX-1 | IDENTIFIED SEQUENCE VARIATIONS |
| Exon 2 | -- | Exon 1 | -- |
| Exon 3 | -- | Intron 1 | -- |
| Exon 4 | SNP rs4904210 G/C | Exon 2 | -- |
| Exon 5 | -- | ||
| CASE 4 | |||
|---|---|---|---|
| PAX-9 | IDENTIFIED SEQUENCE VARIATIONS | MSX-1 | IDENTIFIED SEQUENCE VARIATIONS |
| Exon 2 | -- | Exon 1 | -- |
| Exon 3 | -- | Intron 1 | SNP rs1042484 T/C |
| Exon 4 | -- | Exon 2 | -- |
| Exon 5 | -- | ||
Family n°2
(cases 5-11)
A special mention about the family n° 2 is necessary because, due to a particularly marked ophthalmic phenotype in the youngest child (6 in the reconstruction of the pedigree) that led to more detailed investigations, the diagnostic classification of Axenfeld- Rieger syndrome has been formulated in all family members with the exclusion of the mother.
For this reason, any significant results in the study of cases 5-11 have not been considered valid for the purposes of the research project.
However, in the interests of completeness, results are reported anyway.
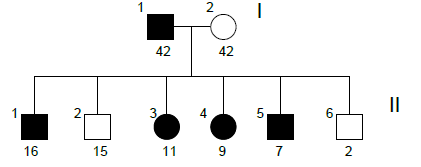
| GRAPHIC IDENTIFICATION | CASE | DATE OF BIRTH | AGENESIS CHARACTERISTICS |
|---|---|---|---|
| I;1 | 5 | 17/01/1969 | 12, 22 |
| I;2 | 6 | 30/01/1969 | non affected |
| II;1 | 7 | 11/05/1994 | 12, 17, 27, 31 |
| II;2 | 9 | 15/12/1995 | non affected |
| II;3 | 8 | 11/09/1999 | 15, 25, 35, 45 |
| II;4 | 10 | 20/07/2001 | 45 |
| II;5 | 11 | 06/07/2003 | 12, 22 |
| II;6 | - | 30/01/2009 | non affected |
Sequencing Results
| CASE 5 | |||
|---|---|---|---|
| PAX-9 | IDENTIFIED SEQUENCE VARIATION | MSX-1 | IDENTIFIED SEQUENCE VARIATION |
| Exon 2 | -- | Exon 1 | -- |
| Exon 3 | -- | Intron 1 | -- |
| Exon 4 | SNP rs4904210 G/C | Exon 2 | -- |
| Exon 5 | -- | ||
Family n° 3
(cases 12-14)
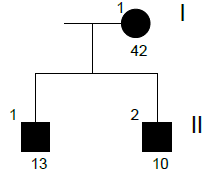
| GRAPHIC IDENTIFICATION | CASE | DATE OF BIRTH | AGENESIS CHARACTERISTICS |
|---|---|---|---|
| I;1 | 12 | 25/09/1968 | 35, 45 |
| II,1 | 14 | 17/03/1998 | 35, 45 |
| II;2 | 13 | 20/04/2000 | 35, 45 |
Sequencing Results
| CASE 12 | |||
|---|---|---|---|
| PAX-9 | IDENTIFIED SEQUENCE VARIATION | MSX-1 | IDENTIFIED SEQUENCE VARIATION |
| Exon 2 | -- | Exon 1 | -- |
| Exon 3 | -- | Intron 1 | SNP rs8670 C/T |
| Exon 4 | -- | Exon 2 | -- |
| Exon 5 | -- | ||
Family n° 4
(cases 15, 16)
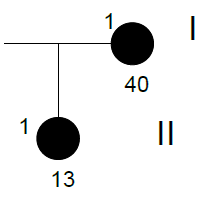
| GRAPHIC IDENTIFICATION | CASE | DATE OF BIRTH | AGENESIS CHARACTERISTICS |
|---|---|---|---|
| I;1 | 15 | 12/03/1971 | 15, 25 |
| II;1 | 16 | 19/12/1997 | 31 |
Sequencing Results
| CASE 15 | |||
|---|---|---|---|
| PAX-9 | IDENTIFIED SEQUENCE VARIATION | MSX-1 | IDENTIFIED SEQUENCE VARIATION |
| Exon 2 | -- | Exon 1 | -- |
| Exon 3 | -- | Intron 1 | -- |
| Exon 4 | -- | Exon 2 | -- |
| Exon 5 | -- | ||
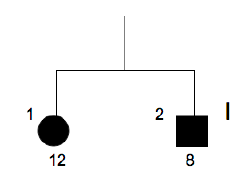
Family n° 5
(cases 17, 18)
| GRAPHIC IDENTIFICATION | CASE | DATE OF BIRTH | IDENTIFIED SEQUENCE VARIATION |
|---|---|---|---|
| I;1 | 18 | 10/10/1999 | 35, 45, 38 e 48 |
| I;2 | 17 | 19/07/2002 | 35, 45 |
Sequencing Results
| CASE 17 | |||
|---|---|---|---|
| PAX-9 | IDENTIFIED SEQUENCE VARIATION | MSX-1 | IDENTIFIED SEQUENCE VARIATION |
| Exon 2 | -- | Exon 1 | -- |
| Exon 3 | -- | Intron 1 | SNP rs1042484 T/C |
| Exon 4 | SNP rs4904210 G/C | Exon 2 | -- |
| Exon 5 | -- | ||
Family n° 6
(cases 19, 20)
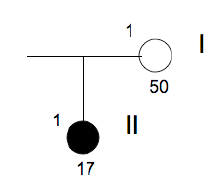
| GRAPHIC IDENTIFICATION | CASE | DATE OF BIRTH |
AGENESIS CHARACTERISTICS |
|---|---|---|---|
| I;1 | 20 | 25/12/1961 | Non affected |
| II;1 | 19 | 08/05/1994 | 15, 17, 18, 25, 28, 34, 35, 38, 44, 45, 46, 48 |
Family n° 7
(cases 21, 22)
| IDENTIFICAZIONE GRAFICA |
CASO | DATA DI NASCITA | CARATTERISTICHE DELL’AGENESIA |
|---|---|---|---|
| I;1 | 21 | 04/03/1971 | Non affected |
| II;1 | 22 | 09/07/1999 | 14,15,17,18,24,25,27,28,32,35,37,38,42,45,47,48 |
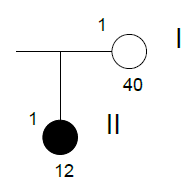
Family n° 8
(cases 23-25)
| GRAPHIC IDENTIFICATION | CASE | DATE OF BIRTH | AGENESIS CHARACTERISTICS |
|---|---|---|---|
| I;1 | 25 | 26/08/1967 | Non affected |
| I;2 | 24 | 06/09/1969 | Non affected |
| II;1 | 23 | 14/01/1998 | 12,22,31,32,41,42 |
Identified Snp
PAX9 – SNP rs4904210 G/C
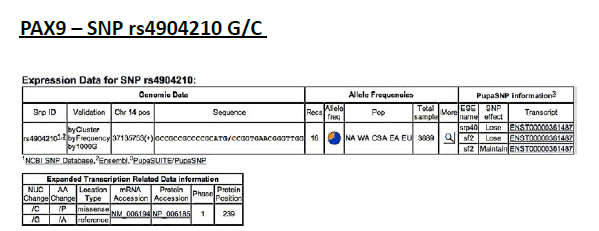
Frequency of minor allele– C: 0,34
ELECTROPHEROGRAM
rs4904210 G>C
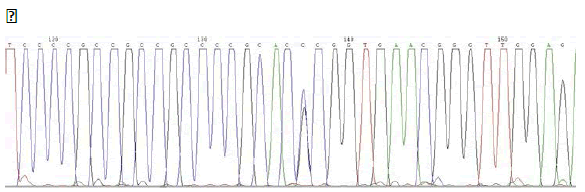
(case n°3)
PAX9 – SNP rs12881240 C/T
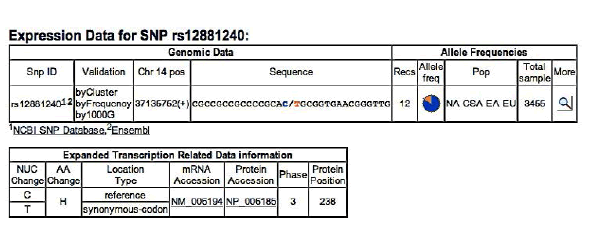
Frequency of minor allel– T: 0,17
ELECTROPHEROGRAM
rs12881240 C>T
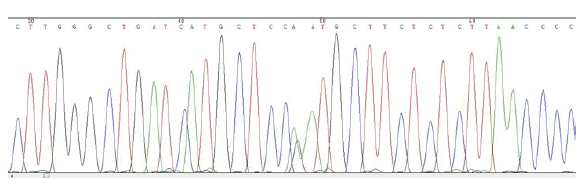
(case n°1)
MSX1 – SNP rs1042484 T/C
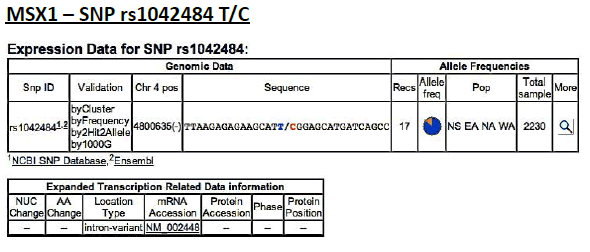
Frequency of minor allele– C: 0,16
ELECTROPHEROGRAM
rs1042484 T>C
reverse sequence
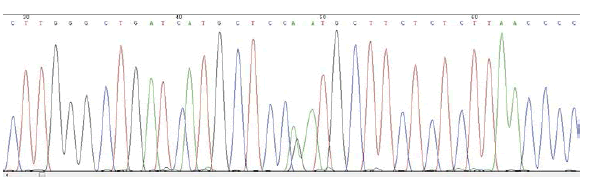
(pilot study)
MSX1 – SNP rs8670 C/T
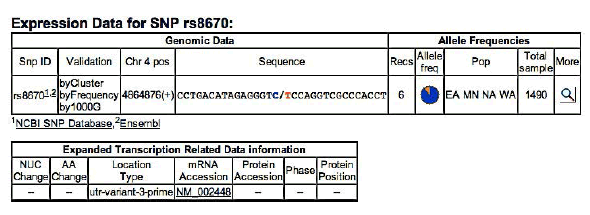
Frequency of minor allele– T: 0,19
ELECTROPHEROGRAM
rs8670 C>T
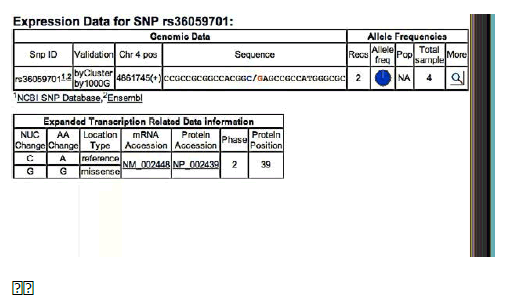
( pilot study)
MSX1 – SNPrs36059701 C/G
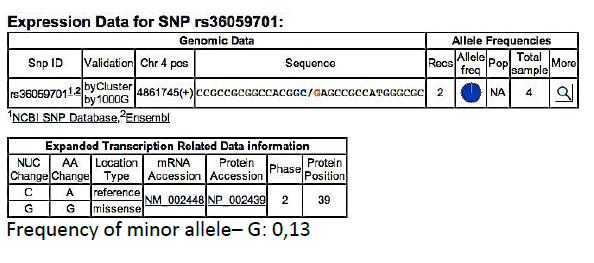
Frequency of minor allele– G: 0,13
ELECTROPHEROGRAM
rs36059701 C>G
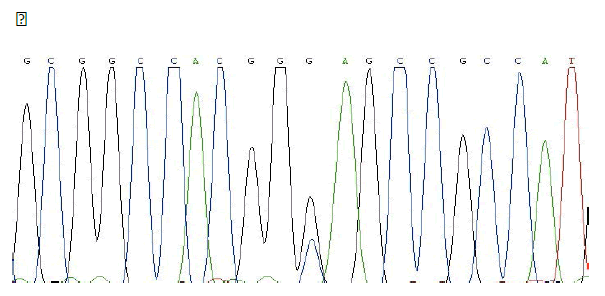
(pilot study)
DISCUSSION
A single nucleotide polymorphism (SNP) is a change of one single nucleotide at a site called the SNP locus. SNPs are the most common type of DNA polymorphism with a frequency of about 1 for 350 bp and constitutes 90-95% of the variations in the DNA sequence.
The new SNP arise spontaneously, such as errors in DNA replication. Since the replication errors are rare, the appearance of new SNP is rare, too.41
The great majority of SNPs happen in non-coding regions of the genome, and in this case, it is called non-coding SNP. The SNPs in the coding regions of the genome, for example in the genes, are called coding SNP (cSNP). Systematic studies of human CSNP have shown that each gene has about 4 cSNP, half of them due to missense mutations in the encoded protein and half due to silent mutations41.
The influence on the phenotype by a CSNP depends on what type of amino acid change of the protein caused the polymorphism. It has been estimated that half of the missense mutations in humans causes genetic diseases.
Genic function may be also influenced by non-coding SNP, when they are located either in the promotor regions or in other regulatory regions41.
Therefore, the detection of SNPs, in particular common or wild type, confirms the data that are now widely accepted by the scientific community that the polymorphisms are sequence variations, which contribute to the development of individual variability of phenotypic traits of each different person.
In fact, in the database dedicated to single nucleotide polymorphisms (dbSNP - https://www.ncbi.nlm.nih.gov/snp/),section of NCBI, National Center for Biotechnology Information, 387 SNP located on gene PAX9 and 197 located on the gene MSX1 have been recorded.
From the obtained results, one can make some considerations:
- Pilot study → only child of non-affected parents. From the sequencing, no mutations in the gene PAX9 are detected, but the presence of three cases of SNPs in the gene MSX1 is found: the first is localized in exon 1 (SNP rs36059701 C / G), the second in intron 1 (SNP rs1042484 T / C) and the third in exon 1 (SNP rs8670 C / T).
- Family n°1→ available withdrawals of mother and three children. Genetic characterization of the mother, case No. 3, has revealed the presence of a SNP (SNP rs4904210 G / C) located in exon 4 of PAX9.
It is interesting to note that this SNP was not inherited by the children (cases n ° 1, 2, 4), in which it is present the same SNP (SNP rs1042484 T / C) located in exon 4 of the gene MSX1. It is assumed that this SNPsia was inherited together with the chromosome set of the father. In the case No. 1 there is another SNP (SNP rs12881240 C / T) located in exon 4 of the gene PAX9.
- Family n°2 →As already mentioned, with regard to all members of the family No. 2, except the mother, Axenfeld- Rieger syndrome was diagnosed, a genetic disease caused by mutations in the genes PITX2 (4q25-q26) or RIEG2 (13q14). Therefore, any significant results would not be considered valid for the purposes of this study. The same sample of study No. 5, the father, was subjected to sequencing, detecting only one SNP (SNP rs4904210 G / C) located in exon 4 of the gene PAX9.
- Family n°3 → pedigree consists of affected mothers and two children. The sequencing of the genes in the case n ° 12, the mother, allowed to confirm the presence of a SNP (SNP rs8670 C / T) in exon 2 of the gene MSX1.
- Family n°4 → pedigree consists of affected mother and daughter, the sample of the mother was analyzed. Any type of gene mutations PAX9 and MSX1 was not detected.
- Family n°5 → Samples of brother and sister, both affected, were collected. The sequencing of the sample belonging to the case n ° 17 has revealed the presence of two SNPs, respectively localized in exon 4 of the gene PAX9 (SNP rs4904210 G / C) and exon 2 of the gene MSX1 (SNP rs1042484 T / C ).
Conclusion
The results cannot be defined as pathognomonic signs of the agenesis. In fact, due to the high diffusion of SNP, it may also be present in individuals without agenesis. In contrast, patients with agenesis may have genes PAX9 and MSX1 without SNP, as demonstrated in this same study by the result of gene sequencing of case No. 15.
In general, the obtained results have allowed us to demonstrate, in agreement with the literature, that odontogenesis is a very complex development process, involving several genetic and molecular factors with specific expression pattern of space-time, which are not yet completely understood.
It is essential to remember that the research focused exclusively on the two genes PAX9 and MSX1. This means that other mutations in different genes possibly involved with a less obvious and more marginal role in the process of morphogenesis dental could be responsible for the agenesis phenotype found in the analyzed cases.
Therefore, wanting to move forward with this type of research on genetic aspects of dental agenesis, the next step will be to use a method of genetic analysis, relatively more modern and efficient, known by the name of ExomeSequencing.
The Exome Sequencing technique, in fact, allows to sequence all the protein-coding genes in a genome (known as the exome), detecting the possible presence of mutations in each gene of the entire DNA of a sample. This method would hypothetically allow to identify new mutations responsible for dental agenesis in those genes that are involved in dental agenesis, even if not as direct as MSX1 and PAX9 or, even, in genes without current evidence of correlation with the development process of dental elements. In addition, this study established an important dialogue between dental clinic and research.
References
- Boeira Junior BR, Echeverrigaray S (2012) Dentistry and molecular biology: a promising field for tooth agenesis management. Tohoku J Exp Med 226: 243-249.
- Valletta G, Bucci E and Matarasso S. (1997) Odontostomatologia. Padova: Piccin.
- Lotito M, Tondo M, Paleotti A. and Negri P. (2009) Stato dell’arte sulle agenesie dentarie: un caso di mancanza del canino superiore. DoctorOs. 20: 1197-1204.
- De Coster PJ, Marks LA, Martens LC, Huysseune A (2009) Dental agenesis: genetic and clinical perspectives. J Oral Pathol Med 38: 1-17.
- Galluccio G, Castellano M, La Monaca C (2012) Genetic basis of non-syndromic anomalies of human tooth number. Arch Oral Biol 57: 918-930.
- Nieminen P (2009) Genetic basis of tooth agenesis. J Exp Zool B Mol Dev Evol 312B: 320-342.
- Lo Muzio L, Mignogna MD, Bucci P, Sorrentino F (1989) Statistical study of the incidence of agenesis in a sample of 1529 subjects. Minerva Stomatol 38: 1045-1051.
- Mattheeuws N, Dermaut L, Martens G (2004) Has hypodontia increased in Caucasians during the 20th century? A meta-analysis. Eur J Orthod 26: 99-103.
- Kirkham J, Kaur R, Stillman EC, Blackwell PG, Elcock C, et al. (2005) The patterning of hypodontia in a group of young adults in Sheffield, UK. Arch Oral Biol 50: 287-291.
- Arte S, Nieminen P, Apajalahti S, Haavikko K, Thesleff I, et al. (2001) Characteristics of incisor-premolar hypodontia in families. J Dent Res 80: 1445-1450.
- Bailleu-Forestier I, Molla M, Verloes A, Berdal A (2008) The genetic basis of inherited anomalies of the teeth. Part 1: clinical and molecular aspects of non-syndromic dental disorders. Eur J Med Genet 51: 273-291.
- Gandolfini M. (1996) Soluzione ortodontica delle agenesie dentarie. Bologna. Edizione Martina.
- Vastardis A (2000) The genetics of human tooth agenesis: new discoveries for understanding dental anomalies. Am J Orthod Dentofacial Orthop 117: 650-656.
- Svinhufvud E, Myllärniemi S, Norio R (1988) Dominant inheritance of tooth malpositions and their association to hypodontia. Clin Genet 34: 373-381.
- Vastardis H, Karimbux N, Guthua SW, Seidman JG, Seidman CE (1996) A human MSX1 homeodomain missense mutation causes selective tooth agenesis. Nat Genet 13: 417-421.
- Schalk-van der Weide Y, Steen WH, Bosman F (1992) Distribution of missing teeth and tooth morphology in patients with oligodontia. ASDC J Dent Child 59: 133-140.
- Cohen MM (1975) Congenital, genetic, and endocrinologic influences on dental occlusion. Dent Clin North Am 19: 499-514.
- van den Boogaard MJ, Dorland M, Beemer FA, van Amstel HK (2000) MSX1 mutation is associated with orofacial clefting and tooth agenesis in humans. Nat Genet 24: 342-343.
- Chishti MS, Muhammad D, Haider M, Ahmad W (2006) A novel missense mutation in MSX1 underlies autosomal recessive oligodontia with associated dental anomalies in Pakistani families. J Hum Genet 51: 872-878.
- Stockton DW, Das P, Goldenberg M, D'Souza RN, Patel PI (2000) Mutation of PAX9 is associated with oligodontia. Nat Genet 24: 18-19.
- Goldenberg M, Das P, Messersmith M, Stockton DW, Patel PI, et al. (2000) Clinical, radiographic, and genetic evaluation of a novel form of autosomal-dominant oligodontia. J Dent Res 79: 1469-1475.
- Russell PJ. (2007) iGenetica, II edizione. EdiSES. Napoli.
Open Access Journals
- Aquaculture & Veterinary Science
- Chemistry & Chemical Sciences
- Clinical Sciences
- Engineering
- General Science
- Genetics & Molecular Biology
- Health Care & Nursing
- Immunology & Microbiology
- Materials Science
- Mathematics & Physics
- Medical Sciences
- Neurology & Psychiatry
- Oncology & Cancer Science
- Pharmaceutical Sciences
